This guest post first appeared on Lukas Novak's (@animalculum) personal website. Guest posts do not necessarily represent the views and policies of ISEP.
The mineral pyrite, iron sulfide gemstone also known as fool’s gold for its gold-like appearance, used to be a favorite material for alchemists in their futile struggles to create precious metals. Since antiquity it is also combined with silver in the so-called marcasite jewelry, popular especially in the 19th century, and until today pyrite is used for production of sulfur dioxide for various industrial applications. Whether you are alchemist, Victorian lady, chemical engineer, or anybody else for that matter, you always have a bit of pyrite with you, or rather inside you. The iron-sulfur (FeS) clusters are molecules composed of iron and sulfur atoms arranged in a pattern resembling pyrite crystal structure, which function as cofactors – small molecular “plug-ins” – in a vital group of proteins involved in such important tasks as electron transfer or DNA repair. FeS clusters are found in all living cells and are indispensable for life as we know it. Their ubiquity and importance even led to formulation of a hypothesis saying that pyrite and similar minerals played a crucial role in the very origin of life.
In eukaryotic cells – building blocks of animals, plants, fungi, and microbial protists – FeS clusters are typically produced by two different metabolic pathways. One set of enzymes (ISC) works in mitochondria. The other (CIA), localized in cytoplasm, provides FeS clusters to all the other parts of the cell including the nucleus. The cytoplasmic pathway doesn’t work on its own, but depends on a, yet unidentified, product of the mitochondrial one. Mitochondria are therefore usually essential for the cell and even their most simplified forms tend to have at least this function intact. And so, production of FeS clusters became a central question for our team after we described the first known eukaryote completely devoid of mitochondria, a chinchilla gut-inhabiting protist Monocercomonoides. How can the cytoplasmic pathway build FeS clusters if mitochondria, together with their pathway, were lost? It turns out the answer is lateral gene transfer – sharing of genetic material between unrelated organisms. The ancestors of Monocercomonoides gained another pathway for FeS cluster synthesis, called SUF, from bacteria and recruited it for work instead of the lost mitochondrial one.
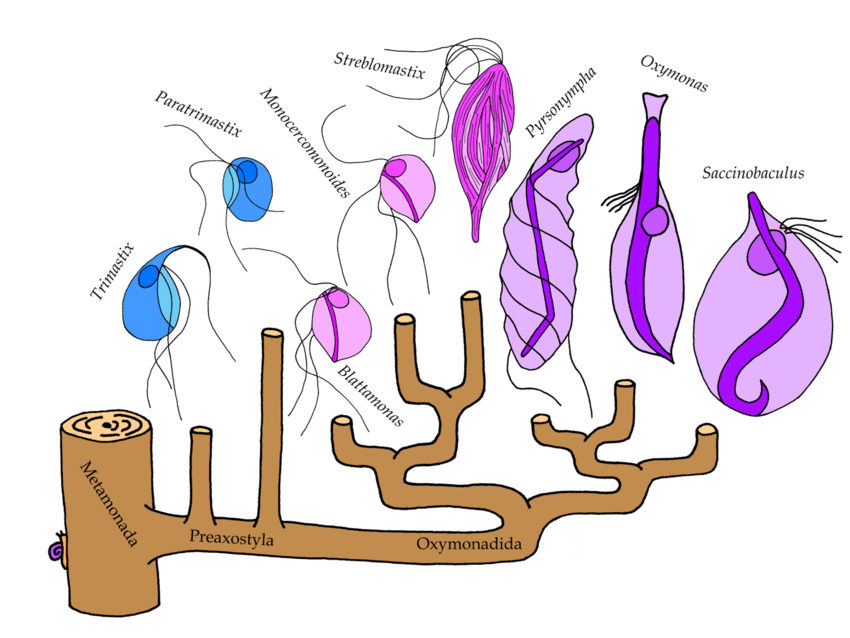
We investigated the evolutionary history of this gene transfer. When did it happen? What other organisms share it? And how did the bacterial SUF pathway change in its new home? We sampled a broad diversity of Monocercomonoides’ relatives constituting a group called Preaxostyla. All of them are single-celled microbes which shun oxygen just like Monocercomonoides, but that’s where the similarity ends. For example, Streblomastixresembles a bundle of snuggly packed symbiotic bacteria, only held together by a network of thin lobes – the actual protist cell. Another one,Saccinobaculus, looks like a bag with a snake inside, which constantly wriggles around. The “snake” is in fact a structure of cellular skeleton that helps the cell to move. Preaxostyla are wonderfully weird creatures indeed! We sequenced 10 species, chosen to cover all major lineages, and found the genes for the SUF pathway in each of them, even those species which, unlike Monocercomonoides, still retain mitochondria. Also, none of the organisms harbored the mitochondrial ISC pathway.
All the 5 genes constituting the SUF pathway in Preaxostyla show the same evolutionary history. That means they must have come in one gene transfer event from bacteria, which happened before all the species split – more than 100 million years in the past. This happened before the mitochondrion vanished, possibly representing the final nail in its coffin. When the mitochondrial pathway was replaced with a new substitute, the microbes simply lost any remaining motivation for keeping the costly organelle and got rid of it. We also found out that 3 of the 5 genes are fused together in Preaxostyla, likely producing a large chimeric protein, a situation not observed in bacteria.
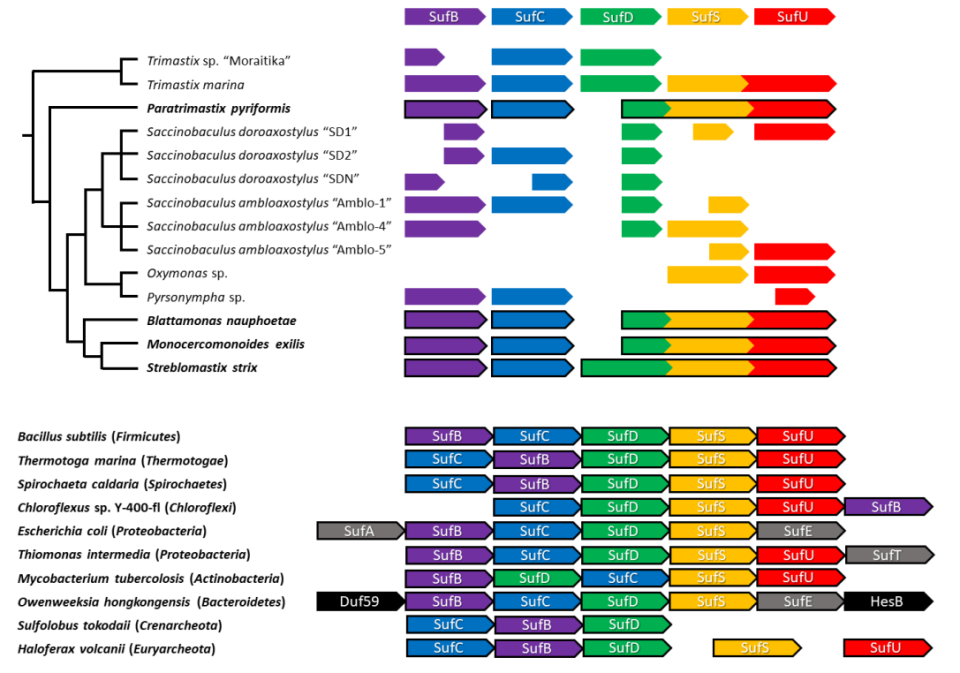
Our findings are most interesting from the evolutionary point of view. They show another strong evidence of lateral gene transfer having a dramatic effect on eukaryotes, a notion which is still controversial. However, they might also have a broader impact in the future. The interesting fusion of 3 genes may indicate that the protein products of these particular genes may be closely cooperating, providing a hint on the general functioning of the SUF pathway. Also, remember that still mysterious connection between mitochondrial and cytoplasmic pathways for FeS cluster production in most eukaryotes including humans? Well, now we have a system where both pathways are in cytoplasm and no mitochondrion is involved. Cross-examination of these different arrangements might help us finally identify the elusive link. Maybe one day, microscopic inhabitants of animal guts will help us uncover the secrets of the gems inside us.
Paper: Vacek V, Novák LVF, Treitli SC, Táborský P, Čepička I, Kolísko M, Keeling PJ, Hampl V: Fe-S Cluster Assembly in Oxymonads and Related Protists.Molecular Biology and Evolution 2018, msy168.


Commentaires